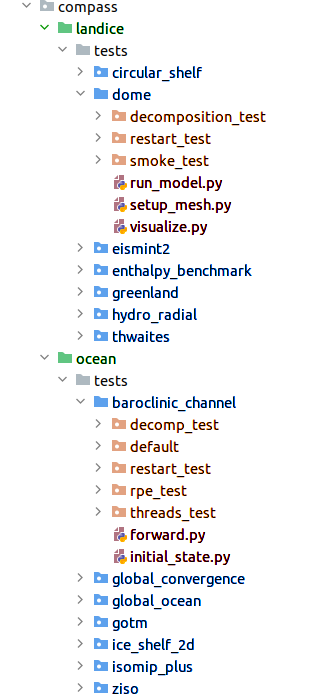
Figure 1: The organization of MPAS cores (green), test groups (blue), test
cases (orange) and steps (red) in the compass package.
Organization of Tests
Here, we describe how tests in compass are organized, both in the package
itself and in the work directories where they get set up and run. At the base
level are MPAS Cores (Landice core or Ocean core). Each MPAS core
has collection of test groups, which has a collection of test cases, each of
which contains a sequence of steps.
Directory structure
In the compass package within a local clone of the compass repository,
MPAS cores, test groups, test cases and steps are laid out like shown in Fig 1.
Each MPAS core has its directory with the compass package directory. Among
other contents of the MPAS core’s directory is a tests directory that
contains all of the test groups. Each test group contains directories for
the test cases and typically also python modules that define the shared steps.
Any steps that are specific to a test case would have a module within that
test case’s directory.
More details on each of these organizational concepts – MPAS Cores, Test Groups, Test cases, and Steps – are provided below.
The organization of the work directory similar but not quite the same as in the
compass package, as shown in Fig. 2.
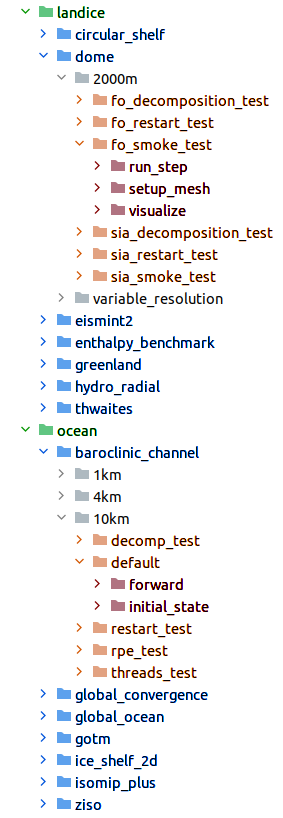
Figure 2: The organization of MPAS cores (green), test groups (blue), test cases (orange) and steps (red) in an example work directory.
At the top level are directories for the MPAS cores. There is no tests
subdirectory in this case – the test groups are directly within the MPAS
core’s directory. The organization of test cases within a test group can
include many additional subdirectories that help sort different versions of
the test cases. In the examples shown above, each test case is in a
subdirectory indicating the resolution of the mesh used in the test case.
Finally, steps are in subdirectories of each test case. In some cases,
additional subdirectories are used to sort steps within a test case (e.g. if
the same step will be run at different mesh resolutions in a convergence test).
MPAS Cores
Currently, there are two MPAS cores, landice, which has test cases for
MALI, and ocean, which encompasses all the test cases for MPAS-Ocean.
From a developer’s perspective, an MPAS core is a package within compass
that has four major pieces:
A class that descends from the
compass.MpasCorebase class. The class is defined in__init__.pyand its__init__()method calls thecompass.MpasCore.add_test_group()method to add each test group to the MPAS core.A
testspackage, which contains packages for each test group, each of which contains various packages and modules for test cases and their steps.An
<mpas_core>.cfgconfig file containing any default config options that are universal to all test groups of the MPAS core.Additional “framework” packages and modules shared between test groups.
The core’s framework is a mix of shared code and other files (config files, namelists, streams files, etc.) that is expected to be used only by modules and packages within the core, not by other cores or the main compass Framework.
The constructor (__init__() method) for a child class of
compass.MpasCore simply calls the parent class’ version
of the constructor with super().__init__(), passing the name of the MPAS
core. Then, it creates objects for each test group and adds them to itself, as
in this example from compass.ocean.Ocean:
from compass.mpas_core import MpasCore
from compass.ocean.tests.baroclinic_channel import BaroclinicChannel
from compass.ocean.tests.global_ocean import GlobalOcean
from compass.ocean.tests.ice_shelf_2d import IceShelf2d
from compass.ocean.tests.ziso import Ziso
class Ocean(MpasCore):
"""
The collection of all test case for the MPAS-Ocean core
"""
def __init__(self):
"""
Construct the collection of MPAS-Ocean test cases
"""
super().__init__(name='ocean')
self.add_test_group(BaroclinicChannel(mpas_core=self))
self.add_test_group(GlobalOcean(mpas_core=self))
self.add_test_group(IceShelf2d(mpas_core=self))
self.add_test_group(Ziso(mpas_core=self))
The object self is always passed to the constructor for each test group
so test groups are aware of which MPAS core they belong to. This is necessary,
for example, in order to create the path for each test group, test case and
step in the work directory.
The config file for the MPAS core should, at the very least, define the
default value for the mpas_model path in the [paths] section. This
path should point to the path within the appropriate E3SM submodule where the
standalone component can be built. This is the path to the directory where the
MPAS component’s executable will be built, not to the executable itself.
Typically, the config file will also define the paths to the model executable and the default namelist and streams files for “forward mode” (and, for the ocean core, “init mode”).
The config file also contains the name of a subdirectory on the
LCRC server
for the dynamical core in the core_path option in the downloads
section:
# This config file has default config options for the landice core
# The paths section points compass to external paths
[paths]
# the relative or absolute path to the root of a branch where MALI has been
# built
mpas_model = MALI-Dev/components/mpas-albany-landice
# The namelists section defines paths to example_compact namelists that will be used
# to generate specific namelists. By default, these point to the forward and
# init namelists in the default_inputs directory after a successful build of
# the landice model. Change these in a custom config file if you need a different
# example_compact.
[namelists]
forward = ${paths:mpas_model}/default_inputs/namelist.landice
# The streams section defines paths to example_compact streams files that will be used
# to generate specific streams files. By default, these point to the forward and
# init streams files in the default_inputs directory after a successful build of
# the landice model. Change these in a custom config file if you need a different
# example_compact.
[streams]
forward = ${paths:mpas_model}/default_inputs/streams.landice
# The executables section defines paths to required executables. These
# executables are provided for use by specific test cases. Most tools that
# compass needs should be in the conda environment, so this is only the path
# to the MALI executable by default.
[executables]
model = ${paths:mpas_model}/landice_model
# Options related to downloading files
[download]
# the path on the server for MALI
core_path = mpas-albany-landice
Test Groups
Test groups are the next level of test-case organization below MPAS Cores. Typically, the test cases within a test group are in some way conceptually linked, serving a similar purpose or being variants on one another. Often, they have a common topography and initial condition, perhaps with different mesh resolutions, parameters, or both. It is common for a test group to include “framework” modules that are shared between its test cases and steps (but not with other test groups). Each MPAS core will typically include a mix of “idealized” test groups (e.g. baroclinic_channel or dome) and “realistic” domains (e.g. greenland and global_ocean).
Each test group is a python package within the core’s tests package.
While it is not required, a test group will typically include a config file,
named <test_group>.cfg, with a set of default config options that are
the starting point for all its test cases. As an example, here is the config
file for the dome test group in the landice core:
# config options for dome test cases
[dome]
# sizes (in cells) for the 2000m uniform mesh
nx = 30
ny = 34
# resolution (in m) for the 2000m uniform mesh
dc = 2000.0
# number of levels in the mesh
levels = 10
# the dome type ('halfar' or 'cism')
dome_type = halfar
# Whether to center the dome in the center of the cell that is closest to the
# center of the domain
put_origin_on_a_cell = True
# whether to add a small shelf to the test
shelf = False
# whether to add hydrology to the initial condition
hydro = False
# config options related to visualization for dome test cases
[dome_viz]
# which time index to visualize
time_slice = 0
# whether to save image files
save_images = True
# whether to hide figures (typically when save_images = True)
hide_figs = True
Some test group options will provide defaults for config options that are
shared across the core (as is the case for the [vertical_grid] config
section in the ocean core). But most config options for a test group will
typically go into a section with the same name as the test group, as in the
example above. Config options that are specific to a particular step might
go into a section with another name, like the [dome_viz] section above.
The __init__.py file for the test group must define a class for the
test group that descends from compass.TestGroup. The constructor
of that class (__init__()) first calls the base class’ constructor with the
parent compass.MpasCore object and the name of the test group.
Then, it constructs objects for each test case in the group and adds them to
itself by calling compass.TestGroup.add_test_case(). Each test case
gets passed the self object as its test group, allowing the test case to
determine both with MPAS core and which test group it belongs to. As an
example, the compass.landice.tests.dome.Dome class looks like this:
from compass.testgroup import TestGroup
from compass.landice.tests.dome.smoke_test import SmokeTest
from compass.landice.tests.dome.decomposition_test import DecompositionTest
from compass.landice.tests.dome.restart_test import RestartTest
class Dome(TestGroup):
"""
A test group for dome test cases
"""
def __init__(self, mpas_core):
"""
mpas_core : compass.landice.Landice
the MPAS core that this test group belongs to
"""
super().__init__(mpas_core=mpas_core, name='dome')
for mesh_type in ['2000m', 'variable_resolution']:
self.add_test_case(
SmokeTest(test_group=self, mesh_type=mesh_type))
self.add_test_case(
DecompositionTest(test_group=self, mesh_type=mesh_type))
self.add_test_case(
RestartTest(test_group=self, mesh_type=mesh_type))
As in this example, it may be useful for a test group to make several
versions of a test case by passing different parameters. In the example, we
create versions of SmokeTest, DecompositionTest and RestartTest
with each of two mesh types (2000m and variable_resolution). We will
explore this further when we talk about Test cases and
Steps below.
It is also common for a test group to define takes care of setting any
additional config options that apply across all test cases but are too
complicated to simply add to the <test_group.cfg> file.
An example of a shared configure() function is
compass.ocean.tests.baroclinic_channel.configure():
def configure(resolution, config):
"""
Modify the configuration options for one of the baroclinic test cases
Parameters
----------
resolution : str
The resolution of the test case
config : configparser.ConfigParser
Configuration options for this test case
"""
res_params = {'10km': {'nx': 16,
'ny': 50,
'dc': 10e3},
'4km': {'nx': 40,
'ny': 126,
'dc': 4e3},
'1km': {'nx': 160,
'ny': 500,
'dc': 1e3}}
if resolution not in res_params:
raise ValueError('Unsupported resolution {}. Supported values are: '
'{}'.format(resolution, list(res_params)))
res_params = res_params[resolution]
for param in res_params:
config.set('baroclinic_channel', param, '{}'.format(res_params[param]))
In the baroclinic_channel test group, 3 resolutions are supported:
1km, 4km and 10km. Here, we use a dictionary to define parameters
(the size of the mesh) associated with each resolution and then to set config
options with those parameters. This approach is appropriate if we want a user
to be able to modify these config options before running the test case (in this
case, if they would like to run on a mesh of a different size or resolution).
If these parameters should be held fixed, they should not be added to the
config object but rather as attributes to the test case’s and/or step’s
class, as we will discuss below.
As with MPAS cores and the main compass package, test groups can also have
a shared “framework” of packages, modules, config files, namelists, and streams
files that is shared among test cases and steps.
Test cases
In many ways, test cases are compass’s fundamental building blocks, since a user can’t set up an individual step of test case (though they can run the steps one at a time).
A test case can be a module but is usually a python package so it can
incorporate modules for its steps and/or config files, namelists, and streams
files. The test case must include a class that descends from
compass.TestCase. In addition to a constructor (__init__()),
the class will often override the configure() and validate() methods of
the base class, as described below.
The run() method in compass.TestCase is deprecated; behaviors
at runtime can instead be handled by individual steps by overriding the
compass.Step.constrain_resources() and
compass.Step.runtime_setup() methods. Details about these methods
are described further in Steps.
TestCase attributes
The base class compass.TestCase has a large number of attributes
that are useful at different stages (init, configuration and run) of the test
case.
Some attributes are available after calling the base class’ constructor
super().__init__(). These include:
self.namethe name of the test case
self.test_groupThe test group the test case belongs to
self.mpas_coreThe MPAS core the test group belongs to
self.subdirthe subdirectory for the test case
self.paththe path within the base work directory of the test case, made up of
mpas_core,test_group, and the test case’ssubdir
Other attributes become useful only after steps have been added to the test case:
self.stepsA dictionary of steps in the test case with step names as keys
self.steps_to_runA list of the steps to run when
compass.run.serial.run_tests()gets called. This list includes all steps by default but can be replaced with a list of only those tests that should run by default if some steps are optional and should be run manually by the user.
Another set of attributes is not useful until configure() is called by the
compass framework:
self.configConfiguration options for this test case, a combination of the defaults for the machine, core and configuration
self.config_filenameThe local name of the config file that
confighas been written to during setup and read from during runself.work_dirThe test case’s work directory, defined during setup as the combination of
base_work_dirandpathself.base_work_dirThe base work directory
These can be used to make further alterations to the config options or to add symlinks files in the test case’s work directory.
Finally, one attribute is available only when the
compass.run.serial.run_tests() function gets called by the
framework:
self.loggerA logger for output from the test case. This gets accessed by other methods and functions that use the logger to write their output to the log file.
You can add other attributes to the child class that keeps track of information
that the test case or its steps will need. As an example,
compass.landice.tests.dome.smoke_test.SmokeTest keeps track of the
mesh type and the velocity solver an attributes:
class SmokeTest(TestCase):
"""
The default test case for the dome test group simply creates the mesh and
initial condition, then performs a short forward run on 4 cores.
Attributes
----------
mesh_type : str
The resolution or type of mesh of the test case
velo_solver : {'sia', 'FO'}
The velocity solver to use for the test case
"""
def __init__(self, test_group, velo_solver, mesh_type):
"""
Create the test case
Parameters
----------
test_group : compass.landice.tests.dome.Dome
The test group that this test case belongs to
velo_solver : {'sia', 'FO'}
The velocity solver to use for the test case
mesh_type : str
The resolution or type of mesh of the test case
"""
name = 'smoke_test'
self.mesh_type = mesh_type
self.velo_solver = velo_solver
subdir = '{}/{}_{}'.format(mesh_type, velo_solver.lower(), name)
super().__init__(test_group=test_group, name=name,
subdir=subdir)
self.add_step(
SetupMesh(test_case=self, mesh_type=mesh_type))
step = RunModel(test_case=self, ntasks=4, openmp_threads=1,
name='run_step', velo_solver=velo_solver,
mesh_type=mesh_type)
if velo_solver == 'sia':
step.add_namelist_options(
{'config_run_duration': "'0200-00-00_00:00:00'"})
self.add_step(step)
step = Visualize(test_case=self, mesh_type=mesh_type)
self.add_step(step, run_by_default=False)
constructor
The __init__() method must first call the base constructor
super().__init__(), passing the name of the test case, the test group it
will belong to, and the subdirectory (if different from the name of the test
case). Then, it should create an object for each step and add them to itself
using call compass.TestCase.add_step().
It is important that __init__() doesn’t perform any time-consuming
calculations, download files, or otherwise use significant resources because
objects get constructed (and all constructors get called) quite often for every
single test case and step in compass: when test cases are listed, set up,
or cleaned up, and also when test suites are set up or cleaned up.
However, it is fine to call the following methods on a step during init because these methods only keep track of a “recipe” for downloading files or constructing namelist and streams files, they don’t actually do the work associated with these steps until the point where the step is being set up in
As an example, here is the constructor from
compass.ocean.tests.baroclinic_channel.rpe_test.RpeTest:
from compass.testcase import TestCase
from compass.ocean.tests.baroclinic_channel.initial_state import InitialState
from compass.ocean.tests.baroclinic_channel.forward import Forward
from compass.ocean.tests.baroclinic_channel.rpe_test.analysis import Analysis
class RpeTest(TestCase):
"""
The reference potential energy (RPE) test case for the baroclinic channel
test group performs a 20-day integration of the model forward in time at
5 different values of the viscosity at the given resolution.
Attributes
----------
resolution : str
The resolution of the test case
"""
def __init__(self, test_group, resolution):
"""
Create the test case
Parameters
----------
test_group : compass.ocean.tests.baroclinic_channel.BaroclinicChannel
The test group that this test case belongs to
resolution : str
The resolution of the test case
"""
name = 'rpe_test'
subdir = f'{resolution}/{name}'
super().__init__(test_group=test_group, name=name,
subdir=subdir)
nus = [1, 5, 10, 20, 200]
res_params = {'1km': {'ntasks': 144, 'min_tasks': 36},
'4km': {'ntasks': 36, 'min_tasks': 8},
'10km': {'ntasks': 8, 'min_tasks': 4}}
if resolution not in res_params:
raise ValueError(
f'Unsupported resolution {resolution}. Supported values are: '
f'{list(res_params)}')
params = res_params[resolution]
self.resolution = resolution
self.add_step(
InitialState(test_case=self, resolution=resolution))
for index, nu in enumerate(nus):
name = 'rpe_test_{}_nu_{}'.format(index + 1, nu)
step = Forward(
test_case=self, name=name, subdir=name,
ntasks=params['ntasks'], min_tasks=params['min_tasks'],
resolution=resolution, nu=float(nu))
step.add_namelist_file(
'compass.ocean.tests.baroclinic_channel.rpe_test',
'namelist.forward')
step.add_streams_file(
'compass.ocean.tests.baroclinic_channel.rpe_test',
'streams.forward')
self.add_step(step)
self.add_step(
Analysis(test_case=self, resolution=resolution, nus=nus))
We have deliberately chosen a fairly complex example to demonstrate how to make full use of Code sharing in a test case.
The test case imports the classes for its steps –
compass.ocean.tests.baroclinic_channel.initial_state.InitialState,
compass.ocean.tests.baroclinic_channel.forward.Forward, and
compass.ocean.tests.baroclinic_channel.rpe_test.analysis.Analysis
– so it can create objects for each and add them to itself with
compass.TestCase.add_step(). After this, the dict of
steps will be available in self.steps.
By default, the test case will go into a subdirectory with the same name as the
test case (rpe_test in this case). However, compass is flexible
about the subdirectory structure and the names of the subdirectories. This
flexibility was an important requirement in moving away from
Legacy COMPASS. Each test case and step must end up in a unique
directory, so it may be important that the name and subdirectory of each test
case or step depends in some way on the arguments passed the constructor. In
the example above, the resolution is an argument to the constructor, which is
then saved as an attribute (self.resolution) and also used to define a
unique subdirectory each resolution: 1km/rpe_test, 4km/rpe_test and
10km/rpe_test.
The same Forward step is included in the test case 5 times with a different
viscosity parameter nu for each. The value of
nu is passed to the step’s constructor, along with
the unique name, subdir, and several other parameters:
resolution, ntasks, and min_tasks. In this example, the steps are
given rather clumsy names – rpe_test_1_nu_1, rpe_test_2_nu_5, etc. –
but these could be any unique names.
configure()
The compass.TestCase.configure() method can be overridden by a
child class to set config options or build them up from defaults stored in
config files within the test case or its test group. The self.config
attribute that is modified in this function will be written to a config file
for the test case (see Config Files).
If you override this method in a test case, you should assume that the
<test_case.name>.cfg file in its package has already been added to the
config options prior to calling configure(). This happens automatically
during test-case setup.
Since many test groups need similar behavior in the configure() method for
each test case, it is common to have a shared function (sometimes also called
configure()) in the test group, as we discussed in Test Groups.
compass.ocean.tests.baroclinic_channel.rpe_test.RpeTest.configure()
simply calls the shared function in its test group,
compass.ocean.tests.baroclinic_channel.configure():
from compass.ocean.tests import baroclinic_channel
def configure(self):
"""
Modify the configuration options for this test case.
"""
baroclinic_channel.configure(self.resolution, self.config)
compass.ocean.tests.baroclinic_channel.configure() was already
shown in Test Groups above. It sets parameters for the number of
cells in the mesh in the x and y directions and the resolution of those cells.
The configure() method can also be used to perform other operations at the
test-case level when a test case is being set up. An example of this would be
creating a symlink to a README file that is shared across the whole test case,
as in compass.ocean.tests.global_ocean.files_for_e3sm.FilesForE3SM.configure():
from compass.io import symlink, package_path
def configure(self):
"""
Modify the configuration options for this test case
"""
self.init.configure(config=self.config)
package = 'compass.ocean.tests.global_ocean.files_for_e3sm'
with package_path(package, 'README') as target:
symlink(str(target), '{}/README'.format(self.work_dir))
The configure() method is not the right place for adding or modifying steps
that belong to a test case. Steps should be added during init and altered only
in their own setup() or runtime_setup() methods.
Test cases that don’t need to change config options don’t need to override
configure() at all.
run()
The functionality of compass.TestCase.run() has been moved to the
compass.run.serial.run_tests() function. The run method is now
deprecated and should not be used to modify runtime processes;
compass.Step.constrain_resources() and
compass.Step.runtime_setup() should be used instead. These methods
are further explained in Steps.
validate()
The base class’s compass.TestCase.validate() can be overridden to
perform Validation of variables in output files from a step and/or
timers from the MPAS model.
In compass.ocean.tests.global_ocean.init.Init.validate(), we see
examples of validation of variables from output files:
def validate(self):
"""
Test cases can override this method to perform validation of variables
and timers
"""
steps = self.steps_to_run
variables = ['temperature', 'salinity', 'layerThickness']
compare_variables(test_case=self, variables=variables,
filename1='initial_state/initial_state.nc')
if self.mesh.with_ice_shelf_cavities:
variables = ['ssh', 'landIcePressure']
compare_variables(test_case=self, variables=variables,
filename1='ssh_adjustment/adjusted_init.nc')
If you leave the default keyword argument skip_if_step_not_run=True,
comparison will be skipped (logging a message) if one or more of the steps
involved in the comparison was not run.
Steps
Steps are the smallest units of work that can be executed on their own in
compass. All test cases are made up of 1 or more steps, and all steps
are set up into subdirectories inside of the work directory for the test case.
Typically, a user will run all steps in a test case but certain test cases may
prefer to have steps that are not run by default (e.g. a long forward
simulation or optional visualization) but which are available for a user to
manually alter and then run on their own.
A step is defined by a class that descends from compass.Step.
The child class must override the constructor and the
compass.Step.run() method, and will sometimes also wish to override
the compass.Step.setup() method, described below.
Step attributes
As was the case for test cases, the base class compass.Step has a
large number of attributes that are useful at different stages (init, setup and
run) of the step.
Some attributes are available after calling the base class’ constructor
super().__init__(). These include:
self.namethe name of the test case
self.test_caseThe test case this step belongs to
self.test_groupThe test group the test case belongs to
self.mpas_coreThe MPAS core the test group belongs to
self.subdirthe subdirectory for the step
self.paththe path within the base work directory of the step, made up of
mpas_core,test_group, the test case’ssubdirand the step’ssubdirself.ntasksthe number of parallel (MPI) tasks the step would ideally use. Too few cores are available on the system to run
ntasks * cpus_per_task, the step will run on all available cores as long as this is not belowmin_tasks * min_cpus_per_taskself.min_tasksthe number of MPI tasks the step requires. If the system fewer than
min_tasks * min_cpus_per_taskcores, the step will failself.cpus_per_taskThe number of CPUs that each task runs with, or the total number of CPUs the step would ideally run with if python threading or multiprocessing is being used, in which case
ntasks = 1self.min_cpus_per_taskThe minimum number of CPUs that each task runs with, or the minimum total number of CPUs required for the step if python threading or multiprocessing is being used, in which case
ntasks = 1. Ifntasks > 1,min_cpus_per_taskmuch be the same ascpus_per_task.self.openmp_threadsthe number of OpenMP threads the step will use
self.cachedWhether to get all of the outputs for the step from the database of cached outputs for the MPAS core that this step belongs to
self.run_as_subprocessWhether to run this step as a subprocess, rather than just running it directly from the test case. It is useful to run a step as a subprocess if there is not a good way to redirect output to a log file (e.g. if the step calls external code that, in turn, calls additional subprocesses).
The default behavior when python code calls one of the
subprocessfunctions is that the output goes tostdout/stderr(i.e. the terminal). When python code outside of compass (e.g.jigsawpy) calls asubprocessfunction (e.g. to call JIGSAW), that output goes to the terminal rather than a log file. For most output tostdout/stderrlikeprint()statements,check_call()in MPAS-Tools employs a “trick” to redirect that output to a log file instead. But that doesn’t work withsubprocesscalls. They continue to go to the terminal. However, if we call a given compass step as a subprocess while redirecting its output to a log file, we can prevent unwanted output from ending up in the terminal (the “outer” subprocess call gets redirected to a log file even when the inner one does not).
Another set of attributes is not useful until setup() is called by the
compass framework:
self.configConfiguration options for this test case, a combination of the defaults for the machine, core and configuration
self.config_filenameThe local name of the config file that
confighas been written to during setup and read from during runself.work_dirThe step’s work directory, defined during setup as the combination of
base_work_dirandpathself.base_work_dirThe base work directory
These can be used to add additional input, output, namelist or streams files based on config options that were not available during init, or which rely on knowing the work directory.
Finally, a few attributes are available only when run() gets called by the
framework:
self.inputsa list of absolute paths of input files produced as part of setting up the step. These input files must all exist at run time or the step will raise an exception
self.outputsa list of absolute paths of output files produced by this step and available as inputs to other test cases and steps. These files must exist after the test has run or an exception will be raised
self.loggerA logger for output from the step. This gets passed on to other methods and functions that use the logger to write their output to the log file.
self.log_filenameThe name of a log file where output/errors from the step are being logged, or
Noneif output is to stdout/stderr
The inputs and outputs should not be altered but they may be used to get file names to read or write.
You can add other attributes to the child class that keeps track of information that the step will need.
As an example,
compass.landice.tests.dome.setup_mesh.SetupMesh keeps track of the
mesh type as an attribute:
from compass.model import make_graph_file
from compass.step import Step
class SetupMesh(Step):
"""
A step for creating a mesh and initial condition for dome test cases
Attributes
----------
mesh_type : str
The resolution or mesh type of the test case
"""
def __init__(self, test_case, mesh_type):
"""
Update the dictionary of step properties
Parameters
----------
test_case : compass.TestCase
The test case this step belongs to
mesh_type : str
The resolution or mesh type of the test case
"""
super().__init__(test_case=test_case, name='setup_mesh')
self.mesh_type = mesh_type
if mesh_type == 'variable_resolution':
# download and link the mesh
# the empty database is a trick for downloading to the root of
# the local MALI file cache
self.add_input_file(filename='mpas_grid.nc',
target='dome_varres_grid.nc', database='')
self.add_output_file(filename='graph.info')
self.add_output_file(filename='landice_grid.nc')
constructor
The step’s constructor (__init__() method) should call the base case’s
constructor with super().__init__(), passing the name of the step, the
test case it belongs to, and possibly several optional arguments: the
subdirectory for the step (if not the same as the name), number of MPI tasks,
the minimum number of MPI tasks, the number of CPUs per task, the minimum
number of CPUs per task, the number of OpenMP threads, and (currently as
placeholders) the amount of memory the step is allowed to use.
Then, the step can add inputs and outputs as well as Adding namelist and streams files, as described below.
As with the test case’s constructor, it is important that the step’s constructor doesn’t perform any time-consuming calculations, download files, or otherwise use significant resources because this function is called quite often for every single test case and step: when test cases are listed, set up, or cleaned up, and also when test suites are set up or cleaned up. However, it is okay to add input, output, streams and namelist files to the step by calling any of the following methods:
Each of these functions just caches information about the the inputs, outputs, namelists or streams files to be read later if the test case in question gets set up, so each takes a negligible amount of time.
The following is from
compass.ocean.tests.baroclinic_channel.forward.Forward():
from compass.step import Step
class Forward(Step):
"""
A step for performing forward MPAS-Ocean runs as part of baroclinic
channel test cases.
Attributes
----------
resolution : str
The resolution of the test case
"""
def __init__(self, test_case, resolution, name='forward', subdir=None,
ntasks=1, min_tasks=None, openmp_threads=1, nu=None):
"""
Create a new test case
Parameters
----------
test_case : compass.TestCase
The test case this step belongs to
resolution : str
The resolution of the test case
name : str
the name of the test case
subdir : str, optional
the subdirectory for the step. The default is ``name``
ntasks : int, optional
the number of tasks the step would ideally use. If fewer tasks
are available on the system, the step will run on all available
tasks as long as this is not below ``min_tasks``
min_tasks : int, optional
the number of tasks the step requires. If the system has fewer
than this number of tasks, the step will fail
openmp_threads : int, optional
the number of OpenMP threads the step will use
nu : float, optional
the viscosity (if different from the default for the test group)
"""
self.resolution = resolution
if min_tasks is None:
min_tasks = ntasks
super().__init__(test_case=test_case, name=name, subdir=subdir,
ntasks=ntasks, min_tasks=min_tasks,
openmp_threads=openmp_threads)
self.add_namelist_file('compass.ocean.tests.baroclinic_channel',
'namelist.forward')
self.add_namelist_file('compass.ocean.tests.baroclinic_channel',
'namelist.{}.forward'.format(resolution))
if nu is not None:
# update the viscosity to the requested value
options = {'config_mom_del2': '{}'.format(nu)}
self.add_namelist_options(options)
# make sure output is double precision
self.add_streams_file('compass.ocean.streams', 'streams.output')
self.add_streams_file('compass.ocean.tests.baroclinic_channel',
'streams.forward')
self.add_input_file(filename='init.nc',
target='../initial_state/ocean.nc')
self.add_input_file(filename='graph.info',
target='../initial_state/culled_graph.info')
self.add_model_as_input()
self.add_output_file(filename='output.nc')
Several parameters are passed into the constructor (with defaults if they
are not included) and then passed on to the base class’ constructor: name,
subdir, ntasks, min_tasks, cpus_per_task,
min_cpus_per_task, and openmp_threads.
Then, two files with modifications to the namelist options are added (for later processing), and an additional config option is set manually via a python dictionary of namelist options.
Then, a file with modifications to the default streams is also added (again, for later processing).
Finally, two input and one output file are added.
constrain_resources()
The constrain_resources() method is used to update the ntasks,
min_tasks, cpus_per_task, and min_cpus_per_task attributes prior to
running the step, in case the user has modified these in the config options.
These performance-related attributes affect how the step runs and must be set
prior to runtime, whereas other options can be set within runtime_setup().
constrain_resources() is called within
compass.run.serial.run_tests(), but can be overridden if desired.
The typical reason to override this function would be to get config options for
ntasks, min_tasks, cpus_per_task, etc. and set the corresponding
attributes. Another reason might be to set these attributes using an algorithm
(e.g. based on the number of cells in the mesh used in the step.)
When overriding constrain_resources, it is important to also call the base
class’ version of the method with super().constrain_resources().
The names of the resources are related to the Slurm naming conventions:
ntasksThe target number of MPI tasks that a step will use if the resources are available.
min_tasksThe minimum number of MPI tasks for a step. If too few resources are available, the step will not run.
cpus_per_taskIf
ntasks > 1, this is typically a number of threads used by each MPI task (e.g. with OpenMP threading). Ifntasks == 1, this may be the number of target cores used in on-node parallelism like python or c++ threading, or python multiprocessing.cpus_per_taskwill automatically be constrained to be less than or equal to the number of cores on a node (and the total available cores). So it may be appropriate to set it to a high value appropriate for machines with large nodes, knowing that it will be constrained to fit on one node.For MPI applications without threading,
cpus_per_taskwill always be1, the default.min_cpus_per_taskThe minimum number of cores for on-node parallelism (threading, multiprocessing, etc.). If too few resources are available, the step will fail with an error message.
setup()
The setup() method is called when a user is setting up the step either
as part of a call to compass setup or compass suite.
As in constructor, you can add input, output, streams and namelist
files to the step by calling any of the following methods:
If you are running the MPAS model, you should call
compass.Step.add_model_as_input() to create a symlink to the
MPAS model’s executable. This can be done in the constructor or the
setup() method.
Set up should not do any major computations or any time-consuming operations
other than downloading files. Time-consuming work should be saved for
run() whenever possible.
As an example, here is
compass.ocean.tests.global_ocean.mesh.mesh.MeshStep.setup():
def setup(self):
"""
Set up the test case in the work directory, including downloading any
dependencies.
"""
# get the these properties from the config options
config = self.config
self.cpus_per_task = config.getint('global_ocean',
'mesh_cpus_per_task')
self.min_cpus_per_task = config.getint('global_ocean',
'mesh_min_cpus_per_task')
Some parts of the mesh computation (creating masks for culling) are done using
python multiprocessing, so the cpus_per_task and min_cpus_per_task
attributes are set to appropriate values based on config options.
runtime_setup()
The runtime_setup() method is used to modify any behaviors of the step at
runtime, in the way that compass.TestCase.run() was previously used.
This includes things like partitioning an MPAS mesh across processors and
computing a times step based on config options that might have been modified
by the user. It must not include modifying the ntasks, min_tasks,
cpus_per_task, min_cpus_per_task or openmp_threads attributes.
These attributes must be altered by overriding
constrain_resources().
run()
Okay, we’re ready to define how the step will run!
The contents of run() can vary quite a lot between steps.
In the baroclinic_channel test group, the run() function for
the initial_state step,
compass.ocean.tests.baroclinic_channel.initial_state.InitialState.run(),
is quite involved:
import xarray
import numpy
from mpas_tools.planar_hex import make_planar_hex_mesh
from mpas_tools.io import write_netcdf
from mpas_tools.mesh.conversion import convert, cull
from compass.ocean.vertical import generate_grid
from compass.step import Step
class InitialState(Step):
...
def run(self):
"""
Run this step of the test case
"""
config = self.config
logger = self.logger
section = config['baroclinic_channel']
nx = section.getint('nx')
ny = section.getint('ny')
dc = section.getfloat('dc')
dsMesh = make_planar_hex_mesh(nx=nx, ny=ny, dc=dc, nonperiodic_x=False,
nonperiodic_y=True)
write_netcdf(dsMesh, 'base_mesh.nc')
dsMesh = cull(dsMesh, logger=logger)
dsMesh = convert(dsMesh, graphInfoFileName='culled_graph.info',
logger=logger)
write_netcdf(dsMesh, 'culled_mesh.nc')
section = config['baroclinic_channel']
use_distances = section.getboolean('use_distances')
gradient_width_dist = section.getfloat('gradient_width_dist')
gradient_width_frac = section.getfloat('gradient_width_frac')
bottom_temperature = section.getfloat('bottom_temperature')
surface_temperature = section.getfloat('surface_temperature')
temperature_difference = section.getfloat('temperature_difference')
salinity = section.getfloat('salinity')
coriolis_parameter = section.getfloat('coriolis_parameter')
ds = dsMesh.copy()
interfaces = generate_grid(config=config)
bottom_depth = interfaces[-1]
vert_levels = len(interfaces) - 1
ds['refBottomDepth'] = ('nVertLevels', interfaces[1:])
ds['refZMid'] = ('nVertLevels', -0.5 * (interfaces[1:] + interfaces[0:-1]))
ds['vertCoordMovementWeights'] = xarray.ones_like(ds.refBottomDepth)
xCell = ds.xCell
yCell = ds.yCell
xMin = xCell.min().values
xMax = xCell.max().values
yMin = yCell.min().values
yMax = yCell.max().values
yMid = 0.5*(yMin + yMax)
xPerturbMin = xMin + 4.0 * (xMax - xMin) / 6.0
xPerturbMax = xMin + 5.0 * (xMax - xMin) / 6.0
if use_distances:
perturbationWidth = gradient_width_dist
else:
perturbationWidth = (yMax - yMin) * gradient_width_frac
yOffset = perturbationWidth * numpy.sin(
6.0 * numpy.pi * (xCell - xMin) / (xMax - xMin))
temp_vert = (bottom_temperature +
(surface_temperature - bottom_temperature) *
((ds.refZMid + bottom_depth) / bottom_depth))
frac = xarray.where(yCell < yMid - yOffset, 1., 0.)
mask = numpy.logical_and(yCell >= yMid - yOffset,
yCell < yMid - yOffset + perturbationWidth)
frac = xarray.where(mask,
1. - (yCell - (yMid - yOffset)) / perturbationWidth,
frac)
temperature = temp_vert - temperature_difference * frac
temperature = temperature.transpose('nCells', 'nVertLevels')
# Determine yOffset for 3rd crest in sin wave
yOffset = 0.5 * perturbationWidth * numpy.sin(
numpy.pi * (xCell - xPerturbMin) / (xPerturbMax - xPerturbMin))
mask = numpy.logical_and(
numpy.logical_and(yCell >= yMid - yOffset - 0.5 * perturbationWidth,
yCell <= yMid - yOffset + 0.5 * perturbationWidth),
numpy.logical_and(xCell >= xPerturbMin,
xCell <= xPerturbMax))
temperature = (temperature +
mask * 0.3 * (1. - ((yCell - (yMid - yOffset)) /
(0.5 * perturbationWidth))))
temperature = temperature.expand_dims(dim='Time', axis=0)
layerThickness = xarray.DataArray(data=interfaces[1:] - interfaces[0:-1],
dims='nVertLevels')
_, layerThickness = xarray.broadcast(xCell, layerThickness)
layerThickness = layerThickness.transpose('nCells', 'nVertLevels')
layerThickness = layerThickness.expand_dims(dim='Time', axis=0)
normalVelocity = xarray.zeros_like(ds.xEdge)
normalVelocity, _ = xarray.broadcast(normalVelocity, ds.refBottomDepth)
normalVelocity = normalVelocity.transpose('nEdges', 'nVertLevels')
normalVelocity = normalVelocity.expand_dims(dim='Time', axis=0)
ds['temperature'] = temperature
ds['salinity'] = salinity * xarray.ones_like(temperature)
ds['normalVelocity'] = normalVelocity
ds['layerThickness'] = layerThickness
ds['restingThickness'] = layerThickness
ds['bottomDepth'] = bottom_depth * xarray.ones_like(xCell)
ds['maxLevelCell'] = vert_levels * xarray.ones_like(xCell, dtype=int)
ds['fCell'] = coriolis_parameter * xarray.ones_like(xCell)
ds['fEdge'] = coriolis_parameter * xarray.ones_like(ds.xEdge)
ds['fVertex'] = coriolis_parameter * xarray.ones_like(ds.xVertex)
write_netcdf(ds, 'ocean.nc')
Without going into all the details of this method, it creates a mesh that
is periodic in x (but not y), then adds a vertical grid and an initial
condition to an xarray.Dataset, which is then written out to
the file ocean.nc.
In the example step we’ve been using,
compass.ocean.tests.baroclinic_channel.forward.Forward.run() looks
like this:
from compass.model import run_model
def run(self):
"""
Run this step of the test case
"""
run_model(self)
the compass.model.run_model() function takes care of updating the
namelist options for the test case to make sure the PIO tasks and stride are
consistent with the requested number of MPI tasks, creates a graph partition
for the requested number of tasks, and runs the model.
To get a feel for different types of run() methods, it may be best to
explore different steps.
inputs and outputs
Currently, steps run in sequence in the order they are added to the test case
(or in the order they appear in the test case’s steps_to_run attribute.
There are plans to allow test cases and their steps to run in parallel in the
future. For this reason, we require that each step defines a list of the
absolute paths to all input files that could come from other steps (possibly in
other test cases) and all outputs from the step that might be used by other
steps (again, possibly in other test cases). There is no harm in including
inputs to the step that do not come from other steps (e.g. files that will be
downloaded when the test case gets set up) as long as they are sure to exist
before the step runs. Likewise, there is no harm in including outputs from the
step that aren’t used by any other steps in any test cases as long as the step
will be sure to generate them.
The inputs and outputs need to be defined during init of either the step or
the test case, or in the step’s setup() method because they are needed
before run() is called (to determine which steps depend on which
other steps). Inputs are added with compass.Step.add_input_file()
and outputs with compass.Step.add_output_file(). Inputs may be
symbolic links to files in compass, from the various databases on the
LCRC server,
downloaded from another source, or from another step.
Because the inputs and outputs need to be defined before the step runs, there can be some cases to avoid. The name of an output file should not depend on a config option. Otherwise, if the user changes the config option, the file actually created may have a different name than expected, in which case the step will fail. This would be true even if a subsequent step would have been able to read in the same config option and modify the name of the expected input file.
Along the same lines, an input or output file name should not depend on data from an input file that does not exist during setup(). Since the file does not exist, there is no way to read the file with the dependency within setup() and determine the resulting input or output file name.
Both of these issues have arisen for the files_for_e3sm test case from the global_ocean test group. Output files are named using the “short name” of the mesh in E3SM, which depends both on config options and on the number of vertical levels, which is read in from a mesh file created in a previous step. For now, the outputs of this step are not used by any other steps so it is safe to simply omit them, but this could become problematic in the future if new steps are added that depend on files_for_e3sm test case.
compass.Step includes several methods for adding input, output,
namelist and streams files:
Input files
Typically, a step will add input files with
compass.Step.add_input_file() during init or in its setup()
method. It is also possible to add inputs in the test case’s
constructor.
It is possible to simply supply the path to an input file as filename
without any other arguments to compass.Step.add_input_file(). In
this case, the file name is either an absolute path or a relative path with
respect to the step’s work directory:
def __init__(self, test_case):
...
self.add_input_file(filename='../setup_mesh/landice_grid.nc')
This is not typically how add_input_file() is used because input files are
usually not directly in the step’s work directory.
Symlinks to input files
The most common type of input file is the output from another step. Rather than just giving the file name directly, as in the example above, the preference is to place a symbolic link in the work directory. This makes it much easier to see if the file is missing (because symlink will show up as broken) and allows you to refer to a short, local name for the file rather than its full path:
import xarray
def __init__(self, test_case):
...
self.add_input_file(filename='landice_grid.nc',
target='../setup_mesh/landice_grid.nc')
...
def run(step, test_suite, config, logger):
...
with xarray.open_dataset('landice_grid.nc') as ds:
...
A symlink is not actually created when compass.Step.add_input_file()
is called. This will not happen until the step gets set up, after calling its
setup() method.
Sometimes you want to create a symlink to an input file in the work directory,
but the relative path between the target and the step’s work directory
isn’t very convenient to determine. This may be because the name of the
subdirectory for this step or the target’s step (or both) depends on
parameters. For such cases, there is a work_dir_target argument that
allows you to give the path with respect to the base work directory (which is
not yet known at init). Here is an example taken from
compass.ocean.tests.global_ocean.forward.ForwardStep:
def __init__(self, test_case, mesh, init, ...):
mesh_path = mesh.mesh_step.path
if mesh.with_ice_shelf_cavities:
initial_state_target = '{}/ssh_adjustment/adjusted_init.nc'.format(
init.path)
else:
initial_state_target = '{}/initial_state/initial_state.nc'.format(
init.path)
self.add_input_file(filename='init.nc',
work_dir_target=initial_state_target)
self.add_input_file(
filename='forcing_data.nc',
work_dir_target='{}/initial_state/init_mode_forcing_data.nc'
''.format(init.path))
self.add_input_file(
filename='graph.info',
work_dir_target='{}/culled_graph.info'.format(mesh_path))
Symlink to input files from compass
Another common need is to symlink a data file from within the test group or test case:
from compass.io import add_input_file
def __init__(self, test_case):
...
self.add_input_file(
filename='enthA_analy_result.mat',
package='compass.landice.tests.enthalpy_benchmark.A')
Here, we supply the name of the package that the file is in. The compass
framework will take care of figuring out where the package is located.
Downloading and symlinking input files
Another type of input file is one that is downloaded and stored locally. Typically, to save ourselves the time of downloading large files and to reduce potential problems on systems with firewalls, we cache the downloaded files in a location where they can be shared between users and reused over time. These “databases” are subdirectories of the core’s database root on the LCRC server.
To add an input file from a database, call
compass.Step.add_input_file() with the database argument:
self.add_input_file(
filename='topography.nc',
target='BedMachineAntarctica_v3_and_GEBCO_2023_0.0125_degree_20230831.nc',
database='bathymetry_database')
In this example from
compass.ocean.tests.global_ocean.init.initial_state.InitialState(),
the file BedMachineAntarctica_v3_and_GEBCO_2023_0.0125_degree_20230831.nc is
slated for later downloaded from
MPAS-Ocean’s bathymetry database.
The file will be stored in the subdirectory mpas-ocean/bathymetry_database
of the path in the database_root config option in the paths section of
the config file. The database_root option is set either by selecting one
of the Supported Machines or in the user’s config file.
You can also specify the database_component parameter to choose to get
files from a database belonging to another component, e.g.:
self.add_input_file(filename='icePresent_QU60km_polar.nc',
target='icePresent_QU60km_polar.nc',
database='partition',
database_component='seaice'
It is also possible to download files directly from a URL and store them in the step’s working directory:
step.add_input_file(
filename='dome_varres_grid.nc',
url='https://web.lcrc.anl.gov/public/e3sm/mpas_standalonedata/'
'mpas-albany-landice/dome_varres_grid.nc')
We recommend against this practice except for very small files.
Copying input files
In nearly all the cases discussed above, a symlink is created to the input
file, usually either from the compass package or from one of the databases.
If you wish to copy the file instead of symlinking it (e.g. so a user can make
local modifications), simply add the keyword argument copy=True to any call
to self.add_input_file():
def __init__(self, test_case):
...
self.add_input_file(filename='landice_grid.nc',
target='../setup_mesh/landice_grid.nc', copy=True)
In this case, a copy of landice_grid.nc will be made in the step’s work
directory.
Output files
We require that all steps provide a list of any output files that other steps are allowed to use as inputs. This helps us keep track of dependencies and will be used in the future to enable steps to run in parallel as long as they don’t depend on each other. Adding an output file is pretty straightforward:
self.add_output_file(filename='output_file.nc')
compass.Step.add_output_file() can be called in a step’s
constructor: or setup() method or (less commonly)
in the test case’s constructor.
The relative path in filename is with respect to the step’s work directory,
and is converted to an absolute path internally before the step is run.
Cached output files
Many compass test cases and steps are expensive enough that it can become
time consuming to run full workflows to produce meshes and initial conditions
in order to test simulations. Therefore, compass provides a mechanism for
caching the outputs of each step in a database so that they can be downloaded
and symlinked rather than being computed each time.
Cached output files are be stored in the compass_cache database within each
MPAS core’s space on that LCRC server (see Downloading and symlinking input files).
If the “cached” version of a step is selected, as we will describe below, each
of the test case’s outputs will have a corresponding “input” file added with
the target being a cache file on the LCRC server and the filename being
the output file. compass uses the cached_files.json database to know
which cache files correspond to which step outputs.
A developer can indicate that compass test suite includes steps with cached
outputs in two ways. First, if all steps in a test case should have cached
output, the following notation should be used:
ocean/global_ocean/QU240/mesh
cached
ocean/global_ocean/QU240/WOA23/init
cached
That is, the word cached should appear after the test case on its own line.
The indentation is for visual clarity and is not required.
Second, ff only some steps in a test case should have cached output, they need to be listed explicitly, as follows:
ocean/global_ocean/QUwISC240/mesh
cached: mesh
ocean/global_ocean/QUwISC240/WOA23/init
cached: initial_state ssh_adjustment
The line can be indented for visual clarity, but must begin with cached:,
followed by a list of steps separated by a single space.
Similarly, a user setting up test cases has two mechanisms for specifying which
test cases and steps should have cached outputs. If all steps in a test case
should have cached outputs, the suffix c can be added to the test number:
compass setup -n 90c 91c 92 ...
In this example, test cases 90 and 91 (mesh and init test cases from
the SOwISC12to30 global ocean mesh, in this case) are set up with cached
outputs in all steps and 92 (performance_test) is not. This approach is
efficient but does not provide any control of which steps use cached outputs
and which do not.
A much more verbose approach is required if some steps use cached outputs and
others do not within a given test case. Each test case must be set up on its
own with the -t and --cached flags as follows:
compass setup -t ocean/global_ocean/QU240/mesh --cached mesh ...
compass setup -t ocean/global_ocean/QU240/WOA23/init --cached initial_state ...
...
Cache files should be generated by first running the test case as normal, then
running the compass cache command-line tool at the base of the work
directory, providing the names of the steps whose outputs should be added to
the cache. The resulting <mpas_core>_cached_files.json should be copied
to compass/<mpas_core>/cached_files.json in a compass branch.
Calls to compass cache must be made on Chrysalis or Anvil. If outputs were
produced on another machine, they must be transferred to one of these two
machines before calling compass cache. File can be added manually to the
LCRC server and the cached_files.json databases but this is not
recommended.
More details on cached outputs are available in the design document Caching outputs from compass steps.
Adding namelist and streams files
MPAS cores, test groups, and test cases can provide namelist and streams files
that are used to replace default namelist options and streams definitions
before MPAS gets run. Namelist and streams files within the compass
package must start with the prefix namelist. and streams.,
respectively, to ensure that they are included when we build the package.
You can make calls to compass.Step.add_namelist_file(),
compass.Step.add_namelist_options() and
compass.Step.add_namelist_file() as described below to indicate how
name list and streams file should be built up by modifying the defaults for the
MPAS model. The namelists and streams files themselves are generated
automatically as part of setting up the test case.
Adding a namelist file
Typically, a step that runs the MPAS model will include one or more calls to
compass.Step.add_namelist_file() within the constructor
or setup() method. Calling this method simply adds the file to
a list that will be parsed if and when the step gets set up. (This way, it is
safe to add namelist files to a step in init even if that test case will never
get set up or run.)
The format of the namelist file is simply a list of namelist options and the replacement values:
config_write_output_on_startup = .false.
config_run_duration = '0000_00:15:00'
config_use_mom_del2 = .true.
config_implicit_bottom_drag_coeff = 1.0e-2
config_use_cvmix_background = .true.
config_cvmix_background_diffusion = 0.0
config_cvmix_background_viscosity = 1.0e-4
Since all MPAS namelist options must have unique names, we do not worry about which specific namelist within the file each belongs to.
A typical namelist file is added by passing a package where the namelist file
is located and the name of the input namelist file within that package
as arguments to compass.Step.add_namelist_file():
self.add_namelist_file('compass.ocean.tests.baroclinic_channel',
'namelist.forward')
If the namelist should have a different name than the default
(namelist.<mpas_core>), the name can be given via the out_name keyword
argument. If init mode is desired, rather than the default, forward
mode, this can also be specified.
Namelist values are replaced by the files (or options, see below) in the sequence they are given. This way, you can add the namelist substitutions for the test group first, and then override those with the replacements for the test case or step.
Adding namelist options
Sometimes, it is easier to replace namelist options using a dictionary within
the code, rather than a namelist file. This is appropriate when there are only
1 or 2 options to replace (so creating a file seems like overkill) or when the
namelist options rely on values that are determined by the code (e.g. different
values for different resolutions). Simply create a dictionary of replacements
and call compass.Step.add_namelist_options() either at init or
in the setup() method of the step. These replacements are parsed, along
with replacements from files, in the order they are added. Thus, you could
add replacements from a namelist file for the test group, test case, or step,
then override them with namelist options in a dictionary for the test case or
step, as in this example:
self.add_namelist_file('compass.ocean.tests.baroclinic_channel',
'namelist.forward')
self.add_namelist_file('compass.ocean.tests.baroclinic_channel',
'namelist.{}.forward'.format(step['resolution']))
if self.nu is not None:
# update the viscosity to the requested value
options = {'config_mom_del2': '{}'.format(step['nu'])}
self.add_namelist_options(options)
Here, we get default options for “forward” steps, then for the resolution of
the test case from namelist files, then update the viscosity nu, which is
an option passed in when creating this step.
Note
Namelist values must be of type str, so use '{}'.format(value) to
convert a numerical value to a string.
Updating namelist options at runtime
It is sometimes useful to update namelist options after a namelist has already
been generated as part of setting up. This typically happens within a step’s
run() method for options that cannot be known beforehand, particularly
options related to the number of MPI tasks, CPUs per task, and OpenMP threads.
In such cases, call compass.Step.update_namelist_at_runtime():
...
replacements = {'config_pio_num_iotasks': '{}'.format(pio_num_iotasks),
'config_pio_stride': '{}'.format(pio_stride)}
self.update_namelist_at_runtime(options=replacements, out_name=namelist)
Adding a streams file
Streams files are a bit more complicated than namelist files because streams files are XML documents, requiring some slightly more sophisticated parsing.
Typically, a step that runs MPAS will include one or more calls to
compass.Step.add_streams_file() within the constructor
or setup() method. Calling this function simply adds the file to
a list within the step dictionary that will be parsed if an when
the step gets set up. (This way, it is safe to add streams files to a step at
init even if that test case will never get set up or run.)
The format of the streams file is essentially the same as the default and generated streams file, e.g.:
<streams>
<immutable_stream name="mesh"
filename_template="init.nc"/>
<immutable_stream name="input"
filename_template="init.nc"/>
<immutable_stream name="restart"/>
<stream name="output"
type="output"
filename_template="output.nc"
output_interval="0000_00:00:01"
clobber_mode="truncate">
<var_struct name="tracers"/>
<var name="xtime"/>
<var name="normalVelocity"/>
<var name="layerThickness"/>
</stream>
</streams>
These are all streams that are already defined in the default forward streams
for MPAS-Ocean, so the defaults will be updated. If only the attributes of
a stream are given, the contents of the stream (the var, var_struct
and var_array tags within the stream) are taken from the defaults. If
any contents are given, as for the output stream in the example above, they
replace the default contents. compass does not include a way to add or
remove contents from the defaults, just keep the default contents or replace
them all. (Legacy COMPASS had such an option but it was found to be mostly
confusing and difficult to keep synchronized with the MPAS code.)
A typical streams file is added by calling
compass.Step.add_streams_file() with a package where the streams
file is located and the name of the input streams file within that package:
self.add_streams_file('compass.ocean.tests.baroclinic_channel',
'streams.forward')
If the streams file should have a different name than the default
(streams.<mpas_core>), the name can be given via the out_name keyword
argument. If init mode is desired, rather than the default, forward
mode, this can also be specified.
Adding a template streams file
The main difference between namelists and streams files is that there is no
direct equivalent for streams of compass.Step.add_namelist_options().
It is simply too confusing to try to define streams within the code.
Instead, compass.Step.add_streams_file() includes a keyword
argument template_replacements. If you provide a dictionary of
replacements to this argument, the input streams file will be treated as a
Jinja2 template that is rendered
using the provided replacements. Here is an example of such a template streams
file:
<streams>
<stream name="output"
output_interval="{{ output_interval }}"/>
<immutable_stream name="restart"
filename_template="../restarts/rst.$Y-$M-$D_$h.$m.$s.nc"
output_interval="{{ restart_interval }}"/>
</streams>
And here is how it would be added, along with replacements:
stream_replacements = {
'output_interval': '00-00-01_00:00:00',
'restart_interval': '00-00-01_00:00:00'}
add_streams_file(step, module, 'streams.template',
template_replacements=stream_replacements)
...
stream_replacements = {
'output_interval': '00-00-01_00:00:00',
'restart_interval': '00-00-01_00:00:00'}
add_streams_file(step, module, 'streams.template',
template_replacements=stream_replacements)
In this example, taken from
compass.ocean.tests.global_ocean.mesh.qu240.dynamic_adjustement.QU240DynamicAdjustment,
we are creating a series of steps that will be used to perform dynamic
adjustment of the ocean model, each of which might have different durations and
restart intervals. Rather than creating a streams file for each step of the
spin up, we reuse the same template with just a few appropriate replacements.
Thus, calls to compass.Step.add_streams_file() with
template_replacements are qualitatively similar to namelist calls to
compass.Step.add_namelist_options().
Updating a streams file at runtime
Just as with namelist options, it is sometimes useful to update streams files
after it has already been generated as part of setting up. This typically
happens within a step’s run() method for properties of the stream that
may be affected by config options that a user may have changed. In such
cases, call compass.Step.update_streams_at_runtime(). In this
fairly complicated example, the duration of the run in hours is a config option
that we turn into a string. A dictionary of replacements together with a
template streams file, as described above, are used to update the streams file
with the new run duration:
import time
from datetime import datetime, timedelta
...
config = self.config
# the duration (hours) of the run
duration = int(3600 * config.getfloat('planar_convergence', 'duration'))
delta = timedelta(seconds=duration)
hours = delta.seconds//3600
minutes = delta.seconds//60 % 60
seconds = delta.seconds % 60
duration = f'{delta.days:03d}_{hours:02d}:{minutes:02d}:{seconds:02d}'
stream_replacements = {'output_interval': duration}
self.update_streams_at_runtime(
'compass.ocean.tests.planar_convergence',
'streams.template', template_replacements=stream_replacements,
out_name='streams.ocean')
Adding MPAS model as an input
If a step involves running MPAS, the model executable can be linked and added
as an input to the step by calling compass.model.add_model_as_input()
in __init__() or the setup() method. This way, if the user has
forgotten to compile the model, this will be obvious by the broken symlink and
the step will immediately fail because of the missing input. The path to the
executable is automatically detected based on the work directory for the step
and the config options.
Test Suites
As described in the Test Suites section of the User’s Guide, COMPASS test cases can be organized into test suites. Each core has separate regression suites, and a core can have multiple independent regression suites. A developer defines a test suite by creating a .txt file within the compass/CORE/suites directory. The format of the .txt file is a list of the work directories to the tests desired to be part of the suite. A line starting with # will be treated as a comment line.