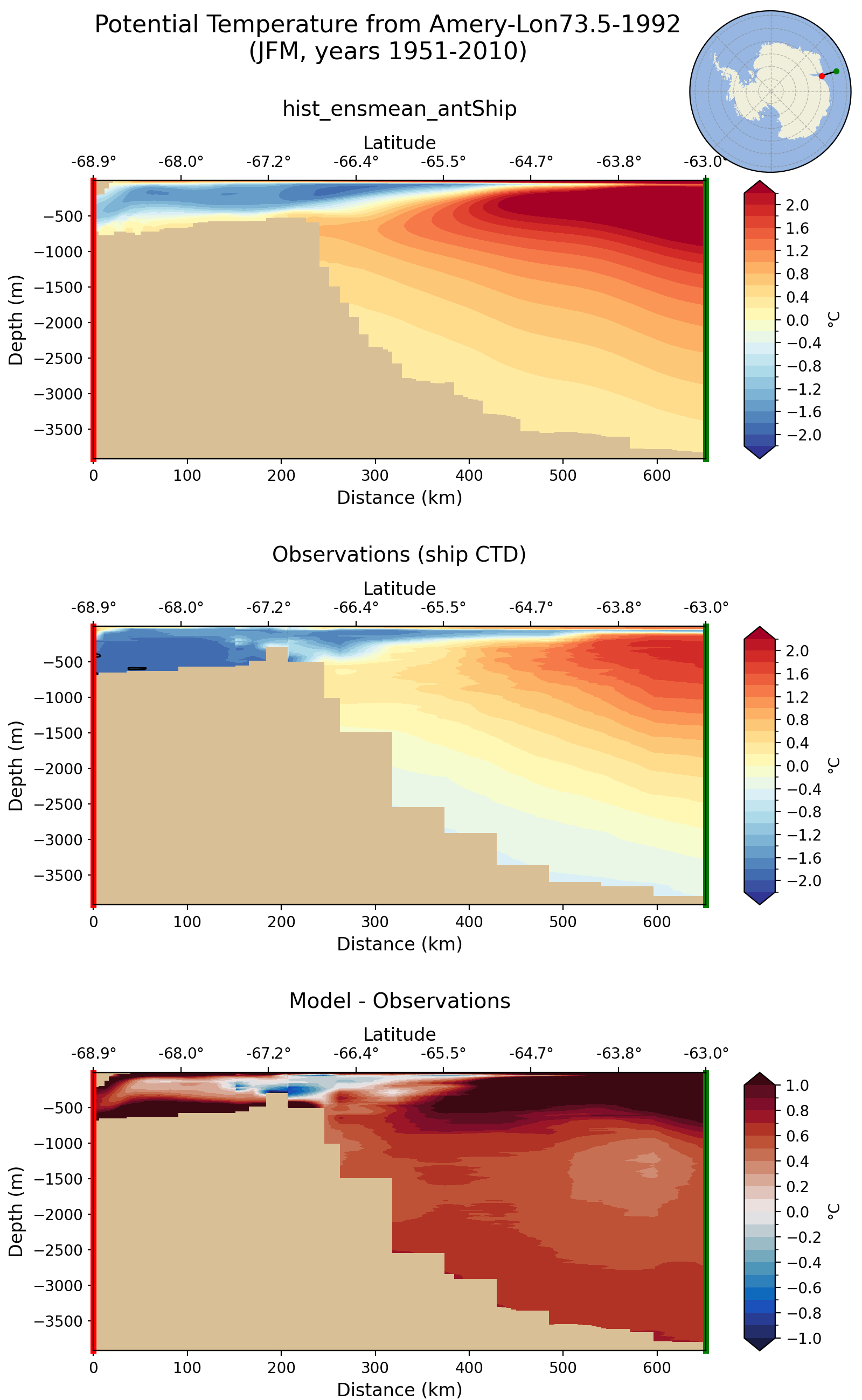
antshipTransects
An analysis task for interpolating MPAS fields to transects from ship-based observations around Antarctica and comparing them.
Component and Tags:
component: ocean
tags: climatology, transect, publicObs
Configuration Options
The following configuration options are available for this task:
[antshipTransects]
## options related to plotting model ship CTD transects.
# Times for comparison times (Jan, Feb, Mar, Apr, May, Jun, Jul, Aug, Sep, Oct,
# Nov, Dec, JFM, AMJ, JAS, OND, ANN)
seasons = ['JFM']
#seasons = ['ANN', 'Jan', 'JFM', 'JAS']
# The approximate horizontal resolution (in km) of each transect. Latitude/
# longitude between observation points will be subsampled at this interval.
# Use 'obs' to indicate no subsampling. Use 'mpas' to indicate plotting of
# model data on the native grid, in which case comparison with observations
# will take place on the observation grid.
#horizontalResolution = mpas
#horizontalResolution = obs
horizontalResolution = 5
# Horizontal bounds of the plot (in km), or an empty list for automatic bounds
# The bounds are a 2-element list of the minimum and maximum distance along the
# transect. Note: A21=Drake Passage; A23=South Atlantic; A12=Prime Meridian
horizontalBounds = {'Belling-BelgicaTrough1-2007': [],
'Belling-BelgicaTrough2-2007': [],
'Belling-Cross1-2007': [],
'Belling-Cross2-2007': [],
'Belling-Cross3-2007': [],
'Belling-EltaninBay-Along-2007': [],
'Belling-GeorgeVI-AlongW1-2007': [],
'Belling-GeorgeVI-AlongW2-2007': [],
'Amundsen-AcrossPIG1-2020': [],
'Amundsen-AcrossPIG2-2020': [],
'Amundsen-AcrossPIG3-2020': [],
'Amundsen-E-Dodson1-1994': [],
'Amundsen-E-Dodson2-1994': [],
'Amundsen-E-PIG-2020': [],
'Amundsen-E-ThwaitesCrosson-1994': [],
'Amundsen-W-PIG-1994': [],
'Amundsen-W-PIG-2009': [],
'Amundsen-W-PIG-2014': [],
'Amundsen-W-PIG-2019': [],
'Ross-C1-2004': [],
'Ross-C2-2004': [],
'Ross-C3-2004': [],
'Ross-C4-2004': [],
'Ross-C5-2004': [],
'Ross-C6-2004': [],
'Ross-C7-2004': [],
'Ross-Shelf1-1994': [],
'Amery-AlongFront-2002': [],
'Amery-AlongFront-2006': [],
'Amery-Lon73.5-1992': [],
'Amery-Lon73.5-2000': [],
'Amery-Lon75.5-1992': [],
'Amery-Lon75.5-2008': [],
'DML-RoiB-E-2008': [],
'DML-RoiB-E-2019': [],
'DML-RoiB-C-1996': [],
'DML-RoiB-C-2006': [],
'DML-RoiB-W-2003': [],
'DML-Munin-Lon18-2003': [],
'DML-Fimbul-E-Lon6.25-1991': [],
'DML-Fimbul-C-Lon0-1992': [],
'DML-Fimbul-C-Lon0-1996': [],
'DML-Fimbul-C-Lon0-2008': [],
'Weddell-RiiserLarsen-C1-1995': [],
'Weddell-RiiserLarsen-C2-1985': [],
'Weddell-RiiserLarsen-C2-2009': [],
'Weddell-RiiserLarsen-C2-2010': [],
'Weddell-RiiserLarsen-C3-1985': [],
'Weddell-RiiserLarsen-C4-1985': [],
'Weddell-RiiserLarsen-C5-1985': [],
'Weddell-RiiserLarsen-C6-1987': [],
'Weddell-StancombBrunt-1990': [],
'Weddell-Filchner-C1-2009': [],
'Weddell-Filchner-C2-2017': [],
'Weddell-Filchner-C3-1995': [],
'Weddell-Filchner-C4-1977': [],
'Weddell-Filchner-C4-1990': [],
'Weddell-Filchner-C5-1978': [],
'Weddell-FilchnerTrough-1995': [],
'Weddell-FilchnerTrough-1999': [],
'Weddell-Ronne-C1-1999': [],
'Weddell-FRISAlongFront-1995': [],
'Weddell-FRISAlongFront-2018': [],
'Weddell-Larsen-C1-1992': [],
'Weddell-Larsen-C2-1992': [],
'Weddell-Larsen-AlongBreak-1992': []}
# The name of the vertical comparison grid. Valid values are 'mpas' for the
# MPAS vertical grid, 'obs' to use the locations of observations or
# any other name if the vertical grid is defined by 'verticalComparisonGrid'.
# If horizontalResolution is 'mpas', model data (both main and control) will be
# plotted on the MPAS vertical grid, regardless of the comparison grid.
#verticalComparisonGridName = mpas
verticalComparisonGridName = obs
#verticalComparisonGridName = uniform_0_to_4000m_at_10m
# The vertical comparison grid if 'verticalComparisonGridName' is not 'mpas' or
# 'obs'. This should be numpy array of (typically negative) elevations (in m).
# The first and last entries are used as axis bounds for 'mpas' and 'obs'
# vertical comparison grids
#verticalComparisonGrid = numpy.linspace(0, -1100, 111)
# A range for the y axis (if any)
verticalBounds = []
# The minimum weight of a destination cell after remapping. Any cell with
# weights lower than this threshold will therefore be masked out.
renormalizationThreshold = 0.01
[antshipTemperatureTransects]
## options related to plotting FRIS transects of potential temperature
# colormap for model/observations
colormapNameResult = RdYlBu_r
# whether the colormap is indexed or continuous
colormapTypeResult = indexed
# color indices into colormapName for filled contours
colormapIndicesResult = numpy.array(numpy.linspace(0, 255, 24), int)
# colormap levels/values for contour boundaries
colorbarLevelsResult = np.arange(-2.2, 2.2 + 0.2, 0.2)
# colormap levels/values for ticks (defaults to same as levels)
colorbarTicksResult = np.arange(-2.0, 2.0 + 0.4, 0.4)
# Adding contour lines to the figure
contourLevelsResult = [-1.9]
contourThicknessResult = 0.5
contourColorResult = black
# colormap for differences
colormapNameDifference = balance
# whether the colormap is indexed or continuous
colormapTypeDifference = indexed
# color indices into colormapName for filled contours
colormapIndicesDifference = numpy.array(numpy.linspace(0, 255, 22), int)
# colormap levels/values for contour boundaries
colorbarLevelsDifference = np.arange(-1.0, 1.0 + 0.1, 0.1)
colorbarTicksDifference = np.arange(-1.0, 1.0 + 0.2, 0.2)
[antshipSalinityTransects]
## options related to plotting FRIS transects of salinity
# colormap for model/observations
colormapNameResult = haline
# whether the colormap is indexed or continuous
colormapTypeResult = indexed
# color indices into colormapName for filled contours
colormapIndicesResult = numpy.array(numpy.linspace(0, 255, 22), int)
# colormap levels/values for contour boundaries
colorbarLevelsResult = np.arange(34.0, 35.0 + 0.05, 0.05)
# colormap levels/values for ticks (defaults to same as levels)
colorbarTicksResult = np.arange(34.0, 35.0 + 0.1, 0.1)
# Adding contour lines to the figure
contourLevelsResult = [34.8]
contourThicknessResult = 0.5
contourColorResult = black
# colormap for differences
colormapNameDifference = balance
# whether the colormap is indexed or continuous
colormapTypeDifference = indexed
# color indices into colormapName for filled contours
colormapIndicesDifference = numpy.array(numpy.linspace(0, 255, 26), int)
# colormap levels/values for contour boundaries
colorbarLevelsDifference = np.arange(-0.6, 0.6 + 0.05, 0.05)
colorbarTicksDifference = np.arange(-0.6, 0.6 + 0.2, 0.2)
[antshipPotentialDensityTransects]
## options related to plotting geojson transects of potential density
# colormap for model/observations
colormapNameResult = Spectral_r
# whether the colormap is indexed or continuous
colormapTypeResult = indexed
# color indices into colormapName for filled contours
colormapIndicesResult = numpy.array(numpy.linspace(0, 255, 22), int)
# colormap levels/values for contour boundaries
colorbarLevelsResult = np.arange(1027.1, 1028.1 + 0.05, 0.05)
# colormap levels/values for ticks (defaults to same as levels)
colorbarTicksResult = np.arange(1027.1, 1028.1 + 0.1, 0.1)
# colormap for differences
colormapNameDifference = balance
# whether the colormap is indexed or continuous
colormapTypeDifference = indexed
# color indices into colormapName for filled contours
colormapIndicesDifference = numpy.array(numpy.linspace(0, 255, 26), int)
# colormap levels/values for contour boundaries
colorbarLevelsDifference = np.arange(-0.6, 0.6 + 0.05, 0.05)
colorbarTicksDifference = np.arange(-0.6, 0.6 + 0.2, 0.2)
[antshipPotentialDensityContourTransects]
## options related to plotting FRIS transects with potential density contours
# Whether to plot the transect as a single contour plot, as opposed to separate
# panels for model, reference and difference.
compareAsContoursOnSinglePlot = True
contourLevelsResult = [1027.2, 1027.4, 1027.6, 1027.7, 1027.8, 1027.85, 1027.9, 1027.95, 1028.0]
- For details on these configuration options, see:
Observations
Example Result