soma
The soma test group (compass.ocean.tests.soma.Soma)
implements variants of the Simulating Ocean Mesoscale Activity (SOMA) test
case (see soma) at resolutions between 4 and 32 km. Here, we
describe the shared framework for this test group and the 5 types of test cases
currently supported: default, long, particles,
surface_restoring, and three_layer.
framework
The shared config options for the soma test group are described
in soma in the User’s Guide.
Additionally, the test group has several shared namelist and streams files,
some for shared parameters and streams for creating the initial condition
(namelist.init and streams.init), some for forward runs
(namelist.forward and streams.forward) and some related to enabling
analysis members in a run and outputting their results (namelist.analysis
and streams.analysis).
initial_state
The class compass.ocean.tests.soma.initial_state.InitialState
defines a step for setting up the initial state for each test case.
First, a mesh appropriate for the resolution is downloaded from the MPAS-Ocean
mesh database. Then, the mesh is culled to remove “land” cells beyond the
1,500 km basin radius. MPAS-Ocean is then run in init mode to generate
the vertical grid, with 100 non-uniform layers according to the
100layerE3SMv1 distribution, and to compute initial temperature and
salinity fields. A constant wind forcing is also generated.
forward
The class compass.ocean.tests.soma.forward.Forward defines a step
for running MPAS-Ocean from the initial condition produced in the
initial_state step. Namelist and streams files are generated when the test
case is set up (combining the *.forward and *.analysis files from the
test group) and updated at runtime based on the config options in the
[soma] section of the config file.
Particles are included in 32km simulations. In
order to partition the particles, we need to first generate the required graph
partition, then partition the particles, and finally run MPAS-Ocean (including
updating PIO namelist options):
cores = self.cores
partition(cores, self.config, self.logger)
if self.with_particles:
section = self.config['soma']
min_den = section.getfloat('min_particle_density')
max_den = section.getfloat('max_particle_density')
nsurf = section.getint('surface_count')
build_particle_simple(
f_grid='mesh.nc', f_name='particles.nc',
f_decomp='graph.info.part.{}'.format(cores),
buoySurf=np.linspace(min_den, max_den, nsurf))
run_model(self, partition_graph=False)
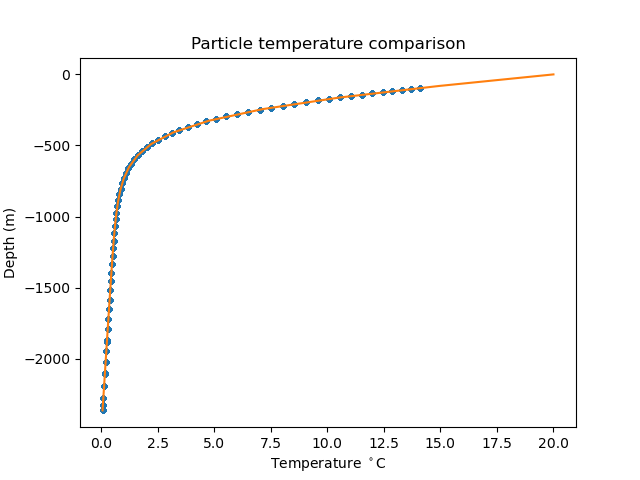
analysis
The class compass.ocean.tests.soma.analysis.Analysis
defines a step for analyzing the results of a forward run with particles
and plotting the particle temperature and salinity against the initial T and S
profiles.
soma_test_case
The compass.ocean.tests.soma.soma_test_case.SomaTestCase class
defines all the SOMA test cases. If a baseline is provided when calling
compass setup, the test case ensures that the final values of
temperature and layerThickness are identical to the baseline values.
If particles are included, a number of particle-related variables and timers
are also validated against the baseline.